Project validation#
datashuttle can validate a project against the
NeuroBlueprint specification.
This will find and display a list of all formatting errors in the project.
To quickly validate an existing project with only the project path, see how to validate a project from a path.
Below we will cover how to validate a datashuttle-managed project (which will additionally log the validation results).
Validating a local project#
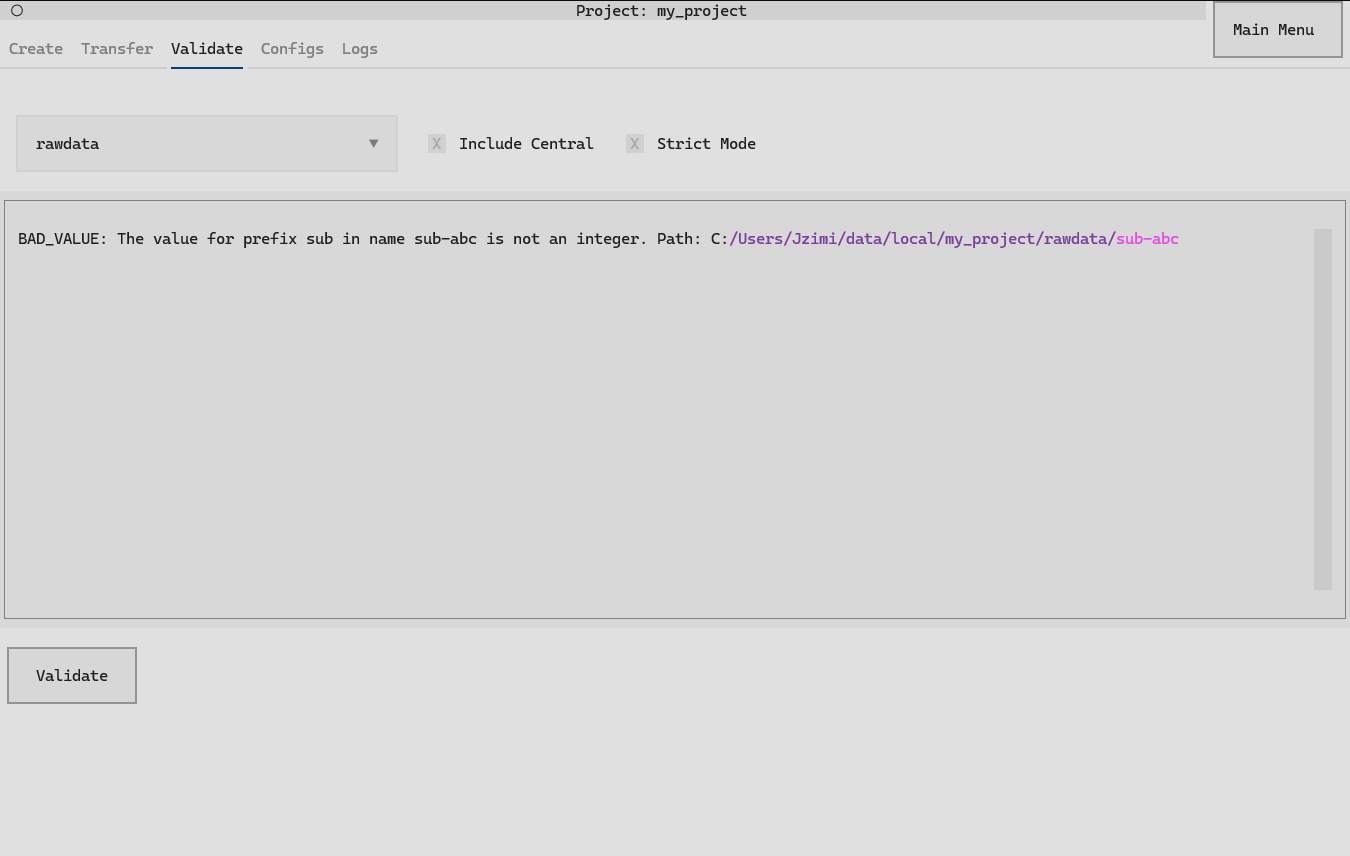
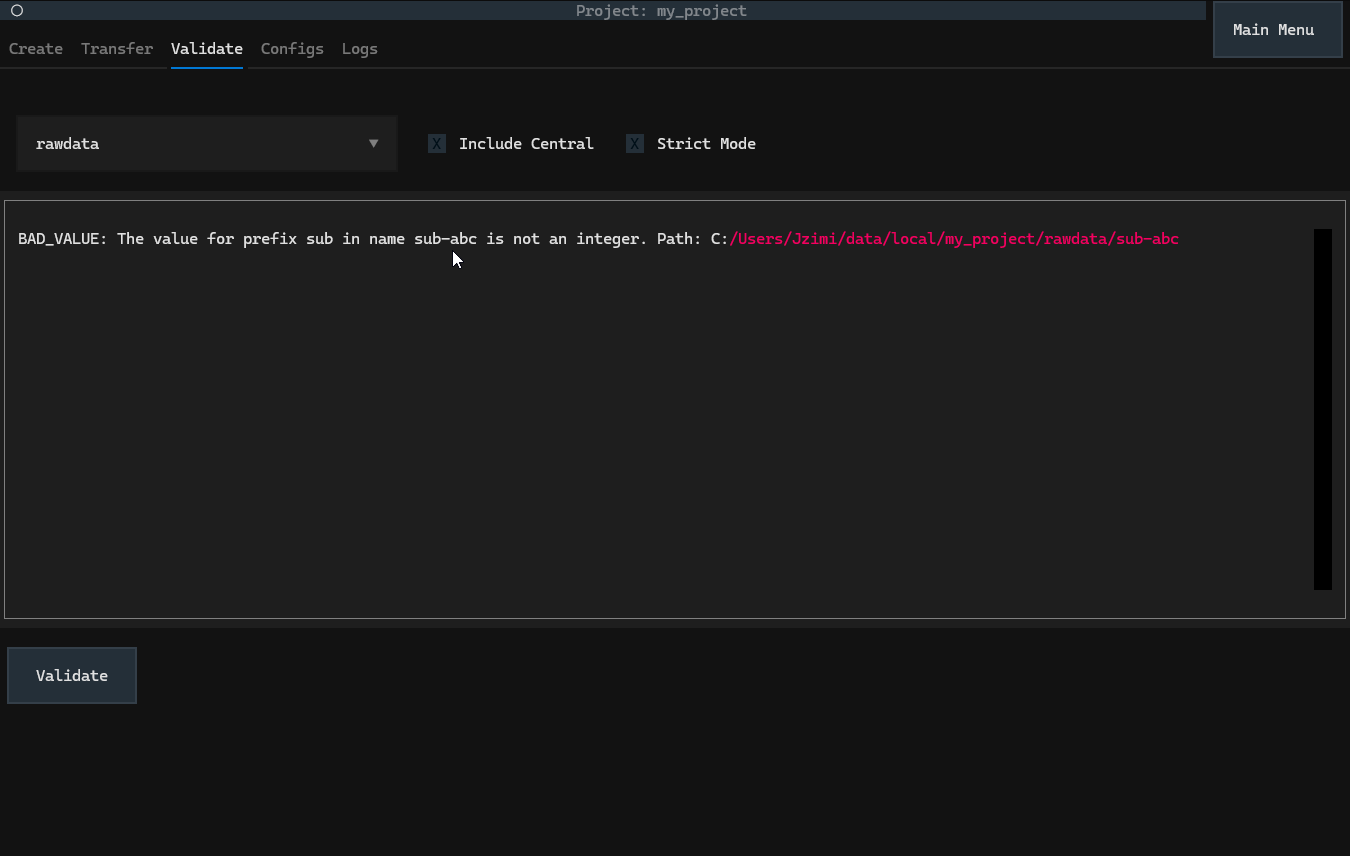
Validation will highlight validation errors within a project. For example, consider
my_project, which has a NeuroBlueprint
error (a subject that does not have an integer value):
└── my_project/
└── rawdata/
└── sub-abc
In a set-up datashuttle project, the Validate tab
can be used to validate the project.
Clicking the validate button will print all validation issues to the output region. See the sections below for details on the options.
Project validation can be run with the validate_project function.
Violations of the NeuroBlueprint can be set to raise an error, be displayed as warnings or printed as output. They are also returned in a list of strings.
from datashuttle import DataShuttle
project = DataShuttle("my_project")
project.make_config_file(local_path="/path/to/my/project") # only required once, on initial project set up
error_messages = project.validate_project(
"rawdata",
display_mode="warn",
)
# UserWarning: BAD_VALUE: The value for prefix sub in name sub-abc is not an integer. Path: <path to folder>
This outputs any NeuroBlueprint validation as a warning.
The returned error_messages is a last of strings containing all validation errors, to be used if required e.g.:
print(error_messages)
# [BAD_VALUE: The value for prefix sub in name sub-abc is not an integer. Path: <path to folder>]
The options for display_mode and "error", "warn" and "print".
For "error", only the first encountered NeuroBlueprint violation will be raised.
Below, we will explore the two key options strict_mode and include_central.
Note
By default, only sub- and ses- prefixed folders are validated
in the project. To validate all folders (including
datatypes)
use strict_mode.
strict_mode#
In strict-mode, all folders outside the datatype folder (e.g. "ephys")
must be NeuroBlueprint-formatted.
NeuroBlueprint does not require all folders in the project to be NeuroBlueprint-formatted sub-, ses- or
datatype folders.
For example, some_other_folder:
└── my_project/
└── rawdata/
├── sub-001/
│ └── ...
└── some_other_folder/
└── ...
However, this means it is hard to validate all folder names, as it is not possible to determine whether
these are mistakes e.g. rat-001 or auxiliary folders. By default, datashuttle will only look for
sub- or ses- prefixed files to validate.
In strict_mode, non-NeuroBlueprint formatted folders are not allowed (except within datatype folders).
Therefore, any additional folders at the subject or session level will raise a validation error, for example:
project.validate_project(
"rawdata",
display_mode="print",
strict_mode=True
)
# BAD_NAME: The name: some_other_folder of type: sub- is not valid. Path: <path to folder>
include_central#
Validation can be performed across all folders in projects in which data is transferred
between a ‘local’ and ‘central’ machine. The validation will combine sub- and ses-
folders across local and central before validation. This is useful check against inconsistent value lengths
(e.g. sub-001 vs sub-02) and duplicate names (e.g. sub-001 and sub-001_date-20240101) across
the local and central project.
To perform this type of validation, connection configurations must be set.
The include_central argument must be set to True:
error_messages = project.validate_project(
"rawdata",
display_mode="warn",
include_central=True
)
allow_letters_in_sub_ses_values#
If True, any alphanumeric character are allowed for the values associated with sub- or ses- keys.
Otherwise, values must be integer and the following additional checks are performed:
Labels must be the same length (e.g.
sub-01andsub-002is invalid).

